Antarctica New Zealand
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Update frequencies
status
-

Data provided here have been collected as part of the project "Measurements and Improved Parameterization of the Thermal Conductivity and Heat Flow through First-Year Sea Ice", OPP-0126007* and include measurements of temperature and various ice properties at selected sites in first-year and multiyear sea ice in McMurdo Sound, Antarctica in the years 2002-2004. Data from earlier installations of thermistor chains for measurements of ice temperature carried out by the New Zealand team have also been included. Data files are in Microsoft Excel format, with individual worksheets for specific cores or temperature data sets. Detailed information and comments on data sampling location etc. are provided in the files. Further information on data collection, results etc. can be found in the following publications: Backstrom, L. G. E., and H. Eicken 2007, submitted, Capacitance probe measurements of brine volume and bulk salinity in first-year sea ice, Cold Reg. Sci. Tech. Pringle, D. J., H. Eicken, H. J. Trodahl, and L. G. E. Backstrom 2007, submitted, Thermal conductivity of landfast Antarctic and Arctic sea ice, J. Geophys. Res. Trodahl, H. J., S. O. F. Wilkinson, M. J. McGuinness, and T. G. Haskell 2001, Thermal conductivity of sea ice; dependence on temperature and depth, Geophys. Res. Lett., 28, 1279-1282. Data are in Microsoft Excel format. Abbreviations: AH = Arrival Heights; CH = Camp Haskell (near Delbridge Islands); VUW = Victoria University Wellington; UAF = University Alaska Fairbanks. RELATED PUBLICATION: https://doi.org/10.1017/jog.2022.108 GET DATA: https://drive.google.com/drive/folders/1ooUH9dPvWT66afFC51Cb0JOHg66rn0sy
-

In Antarctica, ice shelves such as the Ross Ice Shelf (RIS) fringe 75% of the coastline and cover over 1.5 million km2, creating distinct and largely unexplored marine environments. It is fundamental to characterize the communities under these shelves to understand their biogeochemical role and predict how they might respond to future ice-shelf collapse 1,2. While historical studies suggested the RIS harbors active microorganisms 3–5, nothing is known about the composition of these communities. In this study, we profiled the composition, function, and activities of microbial communities in three seawater samples (400, 550, 700 m depth) underlying the shelf interior. We combined rate measurements with multi-omics (i.e. single-cell genomics, metagenomics, metatranscriptomics, and metaproteomics). Overall, below-shelf waters harbour microbial communities of comparable abundance and diversity to deep pelagic waters. Based on the meta-omic data, the community is inferred to be sustained by dark carbon fixation using ammonia, nitrite, and sulfur compounds as electron donors. In turn, these chemolithoautotrophs are predicted to support the aerobic heterotrophic majority and various trophic interactions. Consistently, this study and previous activity measurements suggest that dark carbon fixation is sufficient to sustain prokaryotic heterotrophic production, making the waters below the RIS presumably the largest chemolithotrophic system in the global ocean. Further details are provided at https://doi.org/10.1038/s41467-021-27769-5 GET DATA: https://www.ebi.ac.uk/ena/browser/view/PRJEB35712 GET DATA: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA593264
-

As part of the Scott Base Redevelopment Marine Monitoring Programme, the impact of Scott Base's activities on the local marine environment was assessed. Sampling took place at three sites around Hut Point Peninsula on the southern half of Ross Island during October – November 2019 to assess anthropogenic contamination. Two acoustic doppler current profilers (ADCP; Nortek Signature 500) were deployed, and set with a 2-minute sampling period in 1m vertical depth bins from the seabed to the underside of the ice. Instrument heads were kept ~0.5 m beneath the under-surface. ADCP data were downloaded, extracted from their raw formats, and averaged into 10-minute intervals. A magnetic declination of 141.09° E was applied to the measured current direction to correct the readings to reflect true north and a pressure offset was applied to standardise depths relative to ambient air pressure at the seawater surface. Information on habitats and benthic epifauna assemblage composition were collected using high resolution video across 2 25m transects at ~22m depth. Multiple overlapping passes were made across the seabed transects at ~0.5 m depth contours between ~20 – 26 m in order to create a 2D orthomosaic image of each site. Analysis of the diver-collected video was done using individual frames. The video along each transect was divided into 10 equal time segments and still frames were taken at random from the first, third, fifth, seventh and ninth segments. Eight video frames were analysed per transect (i.e., n=8 per transect and n=16 per site) by one individual to minimise observer bias. Sediment samples were collected by divers to determine contaminant concentrations and sediment characteristics (sediment particle size composition, organic matter content, organic carbon content and algal pigment content) at each site. Sponge species (Sphaerotylus antarcticus and Laternula elliptica) were collected for tissue contaminant analysis. Full description of methods is available at: https://doi.org/10.1007/s00300-023-03181-1 GET DATA: drew.lohrer@niwa.co.nz
-
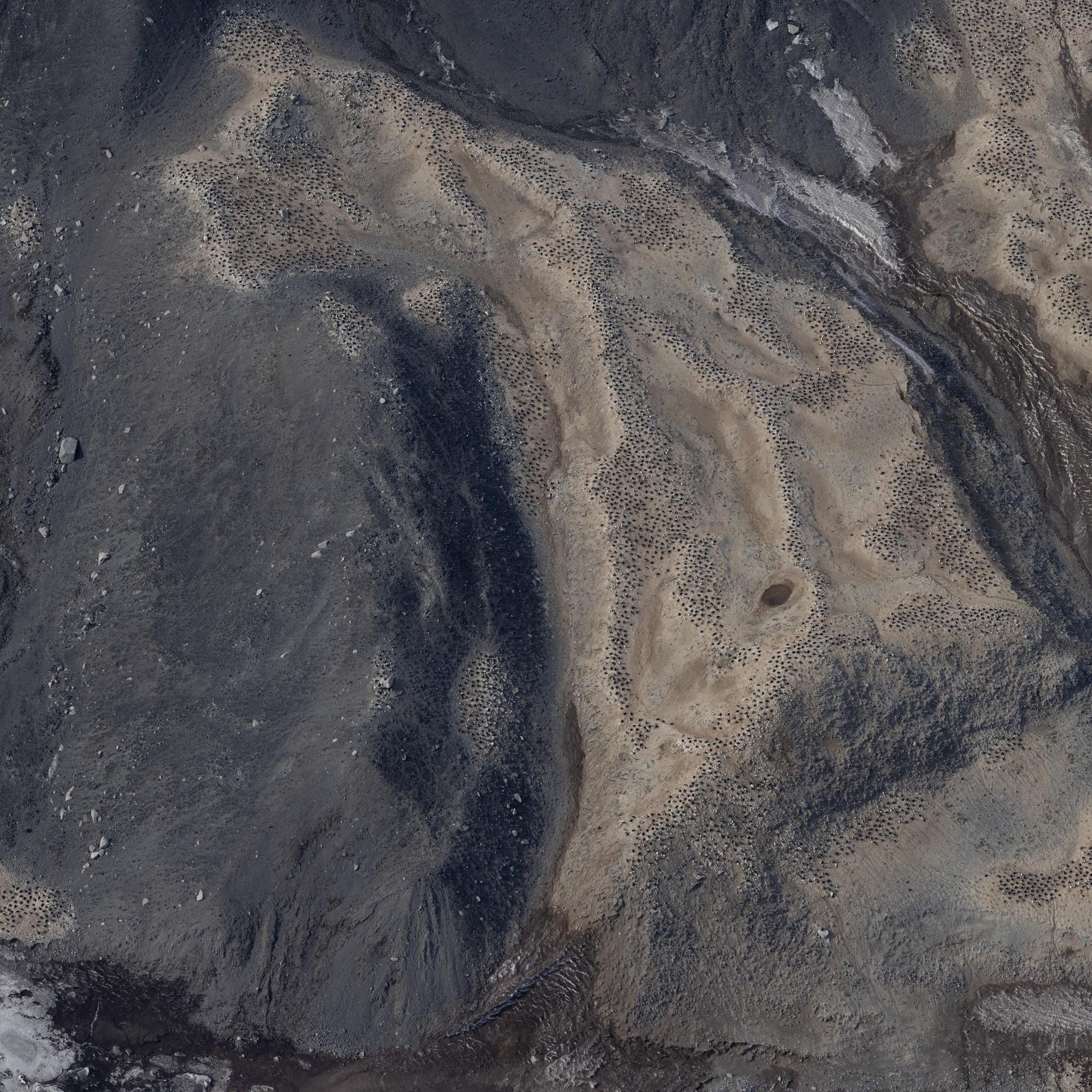
Raw images (over approximately 20.000 unique images) collected during the Adelie Penguin Census across 30+ colonies since 1981 (see associated metadata resource for list full list of colonies). Photographs were taken using a black and white film camera from 1981 to 2004 (inclusive) and 2008. Images are available in .tif fomrat. Images are digital from 2005 onwards (except 2008). Raw images are available in cr2/cr3 formats, and processed images are available across tif and jpg formats. High resolution scanning was initiated in 2011 to scan all the negatives in the collection. There are approximately 10,000 negatives in the collection. Images are taken from helicopter at between 2000-2500 feet. All images collected during the 2324 season are georeferenced with latitude and longitude positions in decimal degrees (WGS 84). Camera Settings: ISO = 400 Shutter speed = greater than 1/1000 Focus = manual, pre-focus to 800m Lens = 135mm with UV filter Aperture/ Exposure = F8 (or up to F11) Image Size = Full size White Balance Setting = Daylight Captures per minute = ~80 GET DATA: m.meredyth-young@antarcticanz.govt.nz
-

Here we present physico-chemical data collected during two research cruises conducted to and across the Ross Sea, Antarctica in the summer of 2018 (February-March) and 2019 (January-February). The dataset includes measurements of temperature, salinity, oxygen, par and transmissivity obtained with a Sea-Bird Electronics (SBE) 911plus CTD. The CTD sensor was configured with SBE 3plus, SBE 4, and SBE 43 dual sensors for the parameters above, in addition to a seapoint fluorescence sensor, and a photosynthetically active radiation (PAR) sensor (Biospherical Instruments QCP‐2300L‐HP). These data were used to provide oceanographic context to DNA metabarcoding analysis of 18S rRNA V4 region that was carried out on DNA samples collected in parallel to nutrient and chlorophyll-a samples. Fastq samples from DNA metabarcoding analysis and the associated metadata (including nutrients, Chlorophyll-a, and size-fractionated chlorophyll-a) were deposited to GenBank under project numbers PRJNA756172 (2018 cruise) and PRJNA974160 (2019 cruise). The study resulting from this analysis has been submitted to Limnology and Oceanography. RELATED PUBLICATION: Cristi, A., Law, C.S., Pinkerton, M., Lopes dos Santos, A., Safi, K. and Gutiérrez-Rodríguez, A. (2024). Environmental driving forces and phytoplankton diversity across the Ross Sea region during a summer–autumn transition. Limnol Oceanogr. https://doi.org/10.1002/lno.12526
-

The Southern Ocean Freshwater Input from Antarctica (SOFIA) Initiative aims to improve our understanding of the simulated response to Antarctic freshwater input, and in particular the model uncertainty. The initiative has developed an experimental protocol and is engaging with modelling groups from around the world to run this common set of experiments using a variety of coupled climate and ocean-only models. Data from this international multi-model ensemble is openly shared for analysis, and is being synthesized into a series of publications by SOFIA participants. Full details are provided at https://doi.org/10.5194/gmd-16-7289-2023 GET DATA: https://sofiamip.github.io/data-access.html
-

This data publication contains biostratigraphic age events for the CIROS-1 drill core, updated age ranges for a suite of samples from the McMurdo erratics sample collection, age-depth tie points for CIROS-1, CRP-2/2A, DSDP 270, DSDP 274, ANDRILL 2A and ANDRILL 1B, and glycerol dialkyl glycerol tetraethers (GDGTs) abundances and indices for samples from the McMurdo erratics, CIROS-1, CRP-2/2A, DSDP 270, DSDP 274, ANDRILL 2A, and ANDRILL 1B. All sample sites are in the Ross Sea region of Antarctica. The McMurdo erratics are glacial erratics collected in the McMurdo Sound region between 1991 and 1996 (Harwood and Levy, 2000). The CIROS-1 drill core was collected from McMurdo sound in 1986 with samples spanning the upper Eocene to lower Miocene. CRP-2/2A drill core was collected in 1999 from offshore Victoria Land with samples for this study from the upper Oligocene-lower Miocene. DSDP Site 270 was recovered from the Eastern Basin of the central Ross Sea in 1973, with samples spanning the upper Oligocene-lower Miocene. DSDP Site 274 was drilled on the lower continental rise in the northwestern Ross Sea in 1973, and samples for this study have been taken from the middle Miocene sections of the drill core. The ANDRILL-2A core was recovered in 2007 from Southern McMurdo Sound, samples span the lower Miocene to middle Miocene and data was originally published in Levy et al. (2016). The ANDRILL-IB core was drilled from the McMurdo Ice Shelf in 2006, samples are compiled from the Plio-Pleistocene section of the core and were originally published in McKay et al. (2012). Biostratigraphic age events are described for CIROS-1, expanding on and updating previously published age models and biostratigraphic ranges. Ages are also revised for the McMurdo erratics by updating the ages of the biostratigraphic markers described by (Harwood and Levy (2000) to more recently published age ranges. Age models for the sample sites are developed using published age datums and the Bayesian age-depth modelling functionality in the R package Bchron (Haslett and Parnell, 2008) to ensure a consistent approach for assigning ages to core depths between datums. GDGT abundances and indices for Ross Sea sites are presented to reconstruct ocean temperatures over the Cenozoic era. Detailed methodology for the processing and analysis of samples for GDGTs is described in the methods section of supplement paper.
-

This metadata record represents the data for generated by mining single-cell genomic, transcriptomic, and metagenomic data to uncover the viral diversity, biogeography, activity, and their role as metabolic facilitators of microbes beneath the Ross Ice Shelf. Hot drilling and seawater sampling was conducted from the sub-shelf water column in the central region of the RIS (Latitude −80.6577 N, Longitude 174.4626 W). The sampling site was located ≈300 km from the shelf front. A borehole (30 cm diameter) conducted by hot water drilling was used for direct sampling of seawater from three depths (400 m, 550 m, and 700 m from the top of the shelf, which correspond to 30 m, 180 m, and 330 m from the bottom of the ice shelf, respectively). Seawater samples were processed accordingly for single cell genomics, metagenomics, and transcriptomics as described5, and the resulting assembled and co-assembled contigs (min. length 1 kb) from single-amplified genomes, bins and transcriptomics were mined for detecting viral contigs. Further details are provided at https://doi.org/10.1038/s41467-023-44028-x GET DATA: https://doi.org/10.6084/m9.figshare.24581331
-

The data is generated through modelling simulations using the University of Victoria Earth system climate model. The modelling dataset presented here corresponds to the study entitled "Transient response of Southern Ocean ecosystems during Heinrich stadials". This dataset contains data files of the complete transient simulations (FW,FE and FWFE) and 40ka-control simulation mentioned in Table 1 and Table 2 of the manuscript. We first performed a control simulation 40ka-control integrating a total of 10000 years. We use only the last 200 years of this control simulation for our analysis. The data is generated through modelling simulations using the University of Victoria Earth system climate model. All the final data is in nc format, which can be easily read by Python/ferret or any other common data analysing software. RELATED PUBLICATION: Saini,H., Meissner,K.J., Menviel,L., & Kvale,K.(2024). Transient response of Southern Ocean ecosystems during Heinrich stadials. Paleoceanography and Paleoclimatology, 39, e2023PA004754. https://doi.org/10.1029/2023PA004754 GET DATA: https://doi.org/10.5061/dryad.k3j9kd5dt
-

This metadata record represents the R phytoclass package. Determine the chlorophyll a (Chl a) biomass of different phytoplankton groups based on their pigment biomarkers. The method uses non-negative matrix factorisation and simulated annealing to minimise error between the observed and estimated values of pigment concentrations (Hayward et al. (2023) https://doi.org/10.1002/lom3.10541). The approach is similar to the widely used 'CHEMTAX' program (Mackey et al. 1996) https://doi.org/10.3354/meps144265), but is more straightforward, accurate, and not reliant on initial guesses for the pigment to Chl a ratios for each phytoplankton group. Further details are provided at: Hayward, A., M. H. Pinkerton, and A. Gutierrez-Rodriguez. 2023. phytoclass: A pigment-based chemotaxonomic method to determine the biomass of phytoplankton classes. Limnol. Oceanogr. Methods 21: 220–241. https://doi.org/10.1002/lom3.10541 GET PACKAGE: https://cran.r-project.org/web/packages/phytoclass/readme/README.html
 GeoData.NZ
GeoData.NZ